[6]:
%matplotlib widget
6) Reviewing IFU results
In this tutorial, we are going to explore some tools to check the measurements from the previous tutorial.
This tutorial can found as a script and a notebook on the examples folder. The observation itself, you will need to download it from the MANGA survey. The galaxy we shall study is SHOC579, a compact galaxy with an intense star forming region.
Let’s start by importing the libraries we need:
Loading the data
[23]:
import numpy as np
import lime
from pathlib import Path
from astropy.io import fits
from astropy.wcs import WCS
from matplotlib import pyplot as plt
Let’s define the inputs and outputs paths
[8]:
# State the data location
cfg_file = Path('../sample_data/manga.toml')
cube_file = Path('../sample_data/spectra/manga-8626-12704-LOGCUBE.fits.gz')
bands_file_0 = Path('../sample_data/SHOC579_MASK0_bands.txt')
spatial_mask_file = Path('../sample_data/SHOC579_mask.fits')
output_lines_log_file = Path('../sample_data/SHOC579_log.fits')
Now, we load the configuration file.
[9]:
# Load the configuration file:
obs_cfg = lime.load_cfg(cfg_file)
# Observation properties
z_obj = obs_cfg['SHOC579']['redshift']
In the 4th tutorial, we opened the .fits file using astropy functions to extract the wavelength and flux arrays, as well as, the WCS. In this ocasion, we are going to use the lime.cube.from_file function to directly read the file into a lime.Cube. In this case we only need to provide the observation redshift:
[10]:
# Load the Cube
shoc579 = lime.Cube.from_file(cube_file, instrument='manga', redshift=z_obj)
/home/vital/PycharmProjects/lime/src/lime/read_fits.py:595: RuntimeWarning: invalid value encountered in sqrt
err_cube = np.sqrt(1 / np.ma.masked_array(ivar_cube, pixel_mask_cube))
WARNING: FITSFixedWarning: PLATEID = 8626 / Current plate
a string value was expected. [astropy.wcs.wcs]
WARNING: FITSFixedWarning: 'datfix' made the change 'Set MJD-OBS to 57277.000000 from DATE-OBS'. [astropy.wcs.wcs]
We can visualize the fittings from the previous tutorial by including the measurements file into the Cube.check.cube lines_file argument
[15]:
shoc579.check.cube('H1_6563A', lines_file=output_lines_log_file, rest_frame=True)
/home/vital/PycharmProjects/lime/venv/lib/python3.11/site-packages/numpy/lib/function_base.py:4737: UserWarning: Warning: 'partition' will ignore the 'mask' of the MaskedArray.
arr.partition(
Additinally, you can extract individual lime.Spectrum from this cube and use the Spectrum.plot functions to review measurements from certain spaxels:
[16]:
# Check the individual spaxel fitting configuration
spaxel = shoc579.get_spectrum(38, 35)
spaxel.load_frame(output_lines_log_file, page='38-35_LINELOG')
spaxel.plot.grid()
You can use  functions to extract and convert indivudal spaxels lines logs from the original .fits file:
functions to extract and convert indivudal spaxels lines logs from the original .fits file:
[17]:
spaxel_log_df = lime.load_frame(output_lines_log_file, page='38-35_LINELOG')
lime.save_frame('../sample_data/38-35_linelog.txt', spaxel_log_df, )
Spatial parameter maps
To take advantage of the IFU data, you need to explore the spatial evolution of these line measurements. You can generate these maps with the lime.save_parameter_maps:
[18]:
# Export the line measurements as spatial maps:
param_list = ['intg_flux', 'intg_flux_err', 'profile_flux', 'profile_flux_err', 'v_r', 'v_r_err']
lines_list = ['H1_4861A', 'H1_6563A', 'O3_4363A', 'O3_4959A', 'O3_5007A', 'S3_6312A', 'S3_9069A', 'S3_9531A']
lime.save_parameter_maps(output_lines_log_file, '../sample_data/', param_list, lines_list,
mask_file=spatial_mask_file, output_file_prefix='SHOC579_', wcs=shoc579.wcs)
[==========] 100% of spaxels from file of spaxels from file (../sample_data/SHOC579_log.fits) read (227 total spaxels)
Please remember: The inputs and outputs of the lime.save_parameter_maps can only be .fits files.
This function produces different outputs depending on its arguments:
The
param_listargument establishes which parameters from the inputoutput_lines_log_filewill be exported into a .fits file. There will be .fits file per item on theparam_list. These files will be stored on theoutput_folderusingoutput_files_prefixand the item name. The item name must follow the parameter notation of the logs measurements.The
line_listarguments establishes, which line measurements are exported to the output parameter .fits files. Each file will have one extension per line.The user can specify a binary mask with the
mask_fileargument. By default default it will use all the masks extensions on the input .fits unless the user specifies certain masks with themask_listargument.
Please remember: It is important to provide an input mask_file argument to make sure only spaxels with scientific data are queried on the input output_lines_log_file.
At this point we plot some flux ratio diagnostics from theses maps. Let’s define them:
[19]:
# State line ratios for the plots
lines_ratio = {'H1': ['H1_6563A', 'H1_4861A'],
'O3': ['O3_5007A', 'O3_4959A'],
'S3': ['S3_9531A', 'S3_9069A']}
Since some of these lines are blended we are going to use the gaussian fluxes
[20]:
# State the parameter map file
fits_file = f'../sample_data/SHOC579_profile_flux.fits'
Let’s loop through these ratios and make the plots:
[24]:
# Loop through the line ratios
for ion, lines in lines_ratio.items():
# Recover the parameter measurements
latex_array = lime.label_decomposition(lines, params_list=['latex_label'])[0]
ratio_map = fits.getdata(fits_file, lines[0]) / fits.getdata(fits_file, lines[1])
Halpha = fits.getdata(fits_file, lines[0])
Hbeta = fits.getdata(fits_file, lines[1])
# Get the astronomical coordinates from one of the headers of the lines log
hdr = fits.getheader(fits_file, lines[0])
wcs_maps = WCS(hdr)
# Create the plot
fig = plt.figure(figsize=(10, 10))
ax = fig.add_subplot(projection=wcs_maps, slices=('x', 'y'))
im = ax.imshow(ratio_map, vmin=np.nanpercentile(ratio_map, 16), vmax=np.nanpercentile(ratio_map, 84))
cbar = fig.colorbar(im, ax=ax)
ax.update({'title': f'SHOC579 flux ratio: {latex_array[0]} / {latex_array[1]}', 'xlabel': r'RA', 'ylabel': r'DEC'})
plt.show()
From these plots we can draw the following conclusions:
The
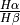 provides a characterisation of the extinction on the SHOC579. This ratio should be above around 2.98, the theoretical emissivity ratio for these lines which is weakly dependant on the physical conditions (temperature and density). However, it seems some pixels have aprubt changes: Can the higher values be only explain by the dust extinction? In the region 1, we didn’t include the wide component of
provides a characterisation of the extinction on the SHOC579. This ratio should be above around 2.98, the theoretical emissivity ratio for these lines which is weakly dependant on the physical conditions (temperature and density). However, it seems some pixels have aprubt changes: Can the higher values be only explain by the dust extinction? In the region 1, we didn’t include the wide component of 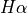 . Does the ratio improve if we include this
profile on the fitting for this region?
. Does the ratio improve if we include this
profile on the fitting for this region?The
![\frac{[OIII]5007Å}{[OIII]4959Å}](../_images/math/3f6ce9d9d9a17edc27d07487b1b585ae13ce9885.png) ratio should remain constant around 3 independently of the physical conditions. In general we observe very close values over the threee masks. What happens if we consider the integrated flux instead of the profile (Gaussian flux). Could it be that this line is too intense and therefore the CCD measurement lies outside the linearity region?
ratio should remain constant around 3 independently of the physical conditions. In general we observe very close values over the threee masks. What happens if we consider the integrated flux instead of the profile (Gaussian flux). Could it be that this line is too intense and therefore the CCD measurement lies outside the linearity region?The
![\frac{[SIII]9531Å}{[SIII]9069Å}](../_images/math/d44b8cf619bcdf5ebf7f12f09a6eb20d993aff33.png) ratio should remain constant around 2.47 independently of the physical conditions. This seems to be the case for most of the galaxy. In some spaxels, however, the value seems to be lower/higher. Could this be explained by the telluric features in the proximity of the lines which could be harder to correct as the emission feature strength becomes weaker? It may be necesarry to inspect these spaxels individually to confirm the if one or both lines are
heavily contaminated.
ratio should remain constant around 2.47 independently of the physical conditions. This seems to be the case for most of the galaxy. In some spaxels, however, the value seems to be lower/higher. Could this be explained by the telluric features in the proximity of the lines which could be harder to correct as the emission feature strength becomes weaker? It may be necesarry to inspect these spaxels individually to confirm the if one or both lines are
heavily contaminated.
As you may have noticed, at this point we are starting to derive scientific conclusions from the data. In any implementation of  , the user is encouraged to review every step to confirm that the measurements agree with the expected results.
, the user is encouraged to review every step to confirm that the measurements agree with the expected results.
[ ]: