[1]:
%matplotlib widget
Spectra
Once you have procured your astronomical observations, the first step in the analysis of spectroscopic lines is opening the files with the actual scientific data. In most cases, this is a .fits file. The reader is recomended to check the official .fits tutorial from astropy.
Not all the astronomical instruments store the data in .fits files in the same way.  has the capability to read .fits files from certain instruments.
has the capability to read .fits files from certain instruments.
In this tutorial, we are going to explore both aproaches: Using  functions and using astropy and numpy to extract the scientific data. The readers are encouraged to contact the author to request that their spectra is added instrument list.
functions and using astropy and numpy to extract the scientific data. The readers are encouraged to contact the author to request that their spectra is added instrument list.
This tutorial can be found as a notebook in the Github examples/inputs folder.
1)  .fits reading functions
.fits reading functions
To check the list of suported instruments use the lime.show_instrument_cfg function:
[2]:
import lime
from pathlib import Path
lime.show_instrument_cfg()
Single ".fits" spectra files configuration:
0 nirspec) units_wave: um, units_flux: MJy, pixel_mask: nan, inst_FWHM: None
1 isis) units_wave: Angstrom, units_flux: FLAM, pixel_mask: nan, inst_FWHM: None
2 osiris) units_wave: Angstrom, units_flux: FLAM, pixel_mask: nan, inst_FWHM: None
3 sdss) units_wave: Angstrom, units_flux: 1e-17*FLAM, pixel_mask: nan, inst_FWHM: None
4 desi) units_wave: Angstrom, units_flux: 1e-17*FLAM, pixel_mask: nan, inst_FWHM: None
Cube ".fits" spectra files configuration:
0 manga) units_wave: Angstrom, units_flux: 1e-17*FLAM,pixel_mask: nan, inst_FWHM: None
1 muse) units_wave: Angstrom, units_flux: 1e-20*FLAM,pixel_mask: nan, inst_FWHM: None
2 megara) units_wave: Angstrom, units_flux: Jy,pixel_mask: nan, inst_FWHM: None
The first list corresponds to the instrument observations, which can be read with the lime.Spectrum.from_file function as longslit spectra, while the second list corresponds to the observations the lime.Cube.from_file loads as IFU cubes. These are some of the parameters from the and the functions respectively.
The previous command also displays the arguments it provides for the `lime.Spectrum <https://lime-stable.readthedocs.io/en/latest/introduction/api.html#lime.Spectrum>`__ and `lime.Cube <https://lime-stable.readthedocs.io/en/latest/introduction/api.html#lime.Cube>`__ functions respectively: * units_wave: Spectrum wavelength units. * units_flux: Spectrum flux units. In some cases, these units include the scale (for example 1e-17for SDSS spectra) with which the flux is
stored. * pixel_mask: Indeces to mask or entries to mask on the wavelength, flux and uncertainty arrays. In the case of pixel_mask=np.nan nan entries in either the flux or the uncertainty flux arrays are masked. * inst_FWHM: Spectrograph FWHM.
Additionally, in the case of and IFU observation, the lime.Cube.from_file function recovers the World Coordidnate System with the observation spatial coordinates.
Please remember: These default configuration values from  may not apply to your observation. This is most likely in observations not comming from surveys. Please remember to confirm these units and configuration parameters match your observations.
may not apply to your observation. This is most likely in observations not comming from surveys. Please remember to confirm these units and configuration parameters match your observations.
Let’s start by reading some of the spectra from the examples/sample_data folder.
[10]:
# Specifying inputs:
data_folder = Path('../sample_data/spectra')
sloan_SHOC579 = data_folder/'sdss_dr18_0358-51818-0504.fits'
nirspec_ceers = data_folder/'hlsp_ceers_jwst_nirspec_nirspec10-001027_comb-mgrat_v0.7_x1d-masked.fits'
Now we can open these files with:
[59]:
spec_shoc579 = lime.Spectrum.from_file(sloan_SHOC579, instrument='sdss', redshift=0.0475)
spec_shoc579.plot.spectrum(rest_frame=True)
[60]:
spec_ceers = lime.Spectrum.from_file(nirspec_ceers, instrument='nirspec', redshift=7.8189, norm_flux=1e-10)
spec_ceers.plot.spectrum(rest_frame=True)
You may notice that in the previous cases we only provided the observation redshift and the norm_flux. The spectra units and pixel masks are already provided by the  functions.
functions.
2) Constructing the  observations from the .fits data
observations from the .fits data
Now, we are going to follow the classical approach, where we extract the scientific data from the .fits files:
a) Long-slit spectra
The calibrated spectrum of a long-slit or echelle instrument consists in a container for the photons dispersion/wavelength, flux and in some cases the flux uncertainty. The latter arrays are usually stored in one of the .fits extensions, however, you might need to reconstruct the wavelength array from the header keys.
OSIRIS instrument at the Gran Telescopio de Canarias
The long-slit spectra from this instruments follows the IRAF standard reduction. Consequently, a page the wavelength array is computed from the CRVAL1, CD1_1 and NAXIS1 keys.
Let’s start by importing the libraries we are going to use and declare the folder with the scientific data:
[61]:
import numpy as np
from astropy.io import fits
from pathlib import Path
from astropy.wcs import WCS
import lime
# Sample data location
data_folder = Path('../sample_data/spectra')
fits_path = data_folder/'gp121903_osiris.fits'
extension = 0
The extension above specifies the page from the .fits file. A quick way to check the extensions in a .fits is the convenience function fits.info:
[62]:
# Fits file information:
fits.info(fits_path)
Filename: ../sample_data/spectra/gp121903_osiris.fits
No. Name Ver Type Cards Dimensions Format
0 PRIMARY 1 PrimaryHDU 143 (3199,) float32
In this file, there is only one extension, so we can now open the file using the astropy.io.fits function:
[63]:
# Open the fits file
with fits.open(fits_path) as hdul:
flux_array, header = hdul[extension].data, hdul[extension].header
In the code above, we use the with context manager. This structure makes sure that the file is closed once we extract the data. This is a safe practice in the case of large .fits files.
Now, we are going to compute the wavelength array:
[64]:
# Reconstruct the wavelength array from the header data
w_min, dw, n_pix = header['CRVAL1'], header['CD1_1'], header['NAXIS1']
w_max = w_min + dw * n_pix
wave_array = np.linspace(w_min, w_max, n_pix, endpoint=False)
print(wave_array)
[ 3626.97753906 3629.04561139 3631.11368372 ... 10236.5367069
10238.60477923 10240.67285156]
In this file the CRVAL1 provides the spectrum lowest wavelength, CD1_1 states the wavelength resolution (constant for this instrument) and the integer NAXIS1 is the number of pixels along the dispersion axis.
Please remember: The keys in a .fits header are not fully standarised. Make sure to check the keys description on the header data and the instrument documentation to read the files correctly.
The units on the spectrum arrays should also be specified on the .fits header:
[65]:
print(f'Wavelength array: {header["WAT1_001"]}')
print(f'Flux array: {header["BUNIT"]}')
Wavelength array: wtype=linear label=Wavelength units=Angstroms
Flux array: erg/cm2/s/A
Now we have can construct a `lime.Spectrum <https://lime-stable.readthedocs.io/en/latest/introduction/api.html#lime.Spectrum>`__:
[66]:
obj = lime.Spectrum(wave_array, flux_array, redshift=0.19531)
obj.plot.spectrum(rest_frame=True)
LiMe INFO: Normalizing input flux by 5.936306664612126e-19
The default units for the `lime.Spectrum <https://lime-stable.readthedocs.io/en/latest/introduction/api.html#lime.Spectrum>`__ and `lime.Cube <https://lime-stable.readthedocs.io/en/latest/introduction/api.html#lime.Cube>`__ are units_wave="Angstrom" and units_flux="FLAM". Since these default values match the ones in these observations we don’t need to provide them.
In the Centimetre–gram–second units system, the flux magnitude is ususally several orders below the unit. Trying to fit a profile at such scales will make the minimizer to fail. Consequently, the user is strongly adviced to provide a norm_flux argument to divide the flux. The output measurents, however, won’t include this normalization.
Please remember: If the observation does not include a norm_flux but the mean spectrum flux value is below 1e-5,  will compute one as in the observation above (notice the terminal message). The user can set
will compute one as in the observation above (notice the terminal message). The user can set norm_flux=1 to avoid the automatic normalization but the fittings will fail if the flux units are very small.
b) SDSS spectrum (data release 18)
The .fits files from SDSS survey are indexed following a dictionary-like structure which includes the wavelength array.
[67]:
sloan_SHOC579 = data_folder/'sdss_dr18_0358-51818-0504.fits'
To load the data we can use:
[68]:
# Open the fits file
extension = 1
with fits.open(sloan_SHOC579) as hdul:
data = hdul[extension].data
header = hdul[extension].header
The flux key indexes the flux. This value is normalized by 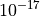 . So we are going to define the units including this scale:
. So we are going to define the units including this scale:
[69]:
flux_array = data['flux']
units_flux = '1e-17*FLAM'
The flux uncertainty is stored as the inversed of the variance. The bad values in this array are set to zero, hence, we are going to mask these values to compute the error spectrum.
[70]:
ivar_array = data['ivar']
err_array = np.sqrt(1/ivar_array)
pixel_mask = ivar_array == 0
/tmp/ipykernel_16108/3510536718.py:2: RuntimeWarning: divide by zero encountered in divide
err_array = np.sqrt(1/ivar_array)
Finally, the wavelength array is stored in logarithmic scale and they are accessed via the loglam key:
[73]:
wave_vac_array = np.power(10, data['loglam'])
This wavelength array is in vacuum. To convert it to air units we can use the Morton (1991, ApJS, 77, 119) law:
[74]:
wave_array = wave_vac_array / (1.0 + 2.735182E-4 + 131.4182 / wave_vac_array**2 + 2.76249E8 / wave_vac_array**4)
Now we can create the Spectrum object:
[76]:
obj = lime.Spectrum(wave_array, flux_array, err_array, pixel_mask=pixel_mask, units_flux=units_flux, redshift=0.0475)
obj.plot.spectrum(rest_frame=True)
/home/vital/PycharmProjects/lime/src/lime/plots.py:738: RuntimeWarning: More than 20 figures have been opened. Figures created through the pyplot interface (`matplotlib.pyplot.figure`) are retained until explicitly closed and may consume too much memory. (To control this warning, see the rcParam `figure.max_open_warning`). Consider using `matplotlib.pyplot.close()`.
fig, ax = plt.subplots()
2) IFU data sets
Integrated field spectrographs provide 3D data array, whose “length” axis corresponds to the dispersion axis while the “height” and “width” axes represent the spatial coordinates. Depending of the instrument, the size of these files can be very large and 3rd party libraries are necessary to open such files.
a) MANGA survey cubes
As we did in the case of the SDSS survey, we are opening an observation of the SHOC579 galaxy, which we are also analysing in the 4th tutorial. Please refer to this tutorial on how to obtain this observation:
[79]:
fits_path = data_folder/'manga-8626-12704-LOGCUBE.fits.gz'
This file is compressed, however, the astropy.io.fits.open can read it. However, keep in mind that reading compressed files takes more time:
[80]:
# Open the MANGA cube fits file
with fits.open(fits_path) as hdul:
# Wavelength 1D array
wave = hdul['WAVE'].data
# Flux 3D array
flux_cube = hdul['FLUX'].data
units_flux = '1e-17*FLAM'
# Convert inverse variance cube to standard error, masking 0-value pixels first
ivar_cube = hdul['IVAR'].data
#pixel_mask_cube = pixel_mask_cube.reshape(ivar_cube.shape)
#err_cube = np.sqrt(1/np.ma.masked_array(ivar_cube, pixel_mask_cube)) * 1e-17
err_cube = np.sqrt(1/ivar_cube)
pixel_mask_cube = ivar_cube == 0
# Header
hdr = hdul['FLUX'].header
/tmp/ipykernel_16108/3592354798.py:15: RuntimeWarning: divide by zero encountered in divide
err_cube = np.sqrt(1/ivar_cube)
The header of .fits spectra usually contains the astronomical coordinates of the observation.  relies on astropy World Coordidnate System function, to plot the IFU data and export the coordinates to the output .fits with your measurements:
relies on astropy World Coordidnate System function, to plot the IFU data and export the coordinates to the output .fits with your measurements:
You can create the WCS object from the corresponding .fits header:
[81]:
# WCS from the observation
wcs = WCS(hdr)
WARNING: FITSFixedWarning: PLATEID = 8626 / Current plate
a string value was expected. [astropy.wcs.wcs]
WARNING: FITSFixedWarning: 'datfix' made the change 'Set MJD-OBS to 57277.000000 from DATE-OBS'. [astropy.wcs.wcs]
Now, we have all the create our Cube object:
[82]:
cube = lime.Cube(wave, flux_cube, err_cube, redshift=0.0475, units_flux=units_flux, pixel_mask=pixel_mask_cube)
Now you can extract individual spaxels to analyse the object lines. Each spaxel will have its corresponding error spectrum and pixel mask
[83]:
# Extract spaxel
spaxel = cube.get_spectrum(38, 35)
# Plot spectrum
spaxel.plot.spectrum(rest_frame=True)
Finally, we can perform measurements on this spaxel using the Spectrum attributes
[84]:
# Fit emission line using the bands from the default database
spaxel.fit.bands('S3_6312A')
# Plot last fitting
spaxel.plot.bands()