[1]:
%matplotlib widget
Line Bands
The second input in a  measurement is the line bands. These are the intervales with the line location and two adjacent and featureless continua. This design was inspired by the the Lick indexes by Worthey et al (1993) and references therein.
measurement is the line bands. These are the intervales with the line location and two adjacent and featureless continua. This design was inspired by the the Lick indexes by Worthey et al (1993) and references therein.
This tutorial can be found as a notebook in the Github examples/inputs folder.
Design
The image below shows an example of the bands for the ![[NeIII]3869Å](../_images/math/4a1fc9a0f6a7f188853b4f26e2baffd38233d9a8.png) line:
line:
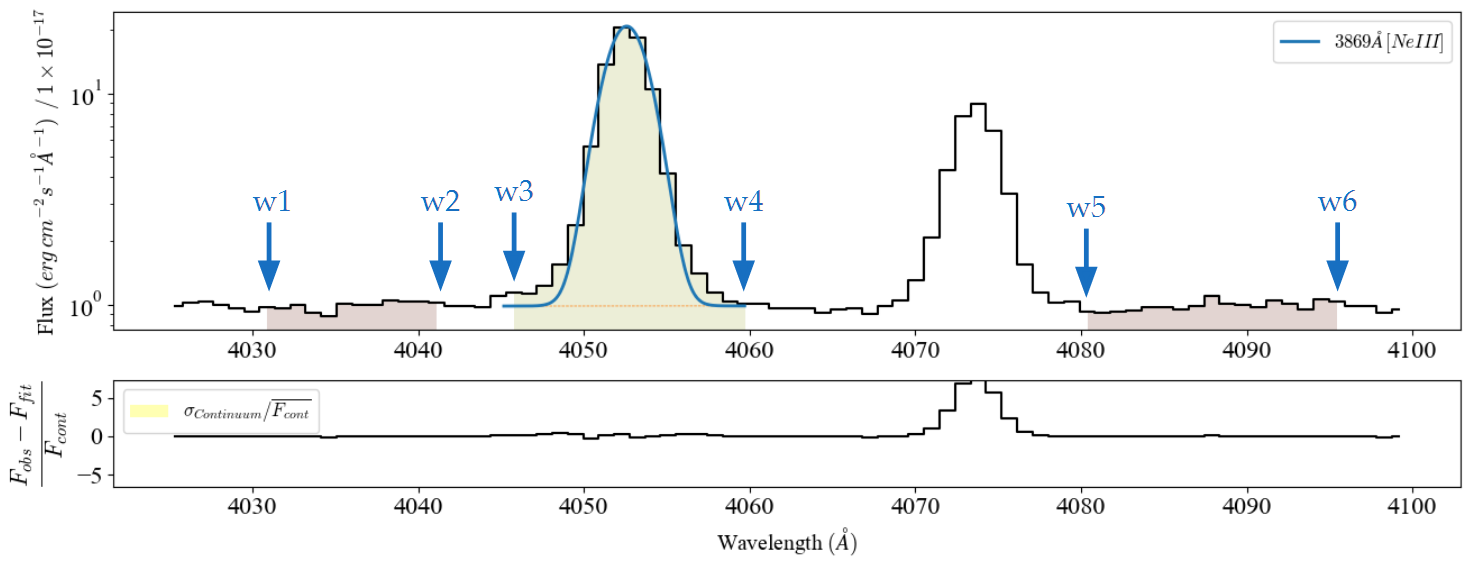
A band consists in a 6-value array 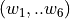 with the wavelength boundaries for the line location and two adjacent continua. For
with the wavelength boundaries for the line location and two adjacent continua. For  measurements, it is essential that:
measurements, it is essential that:
The wavelenght array is sorted from lower to higher values.
The wavelength values are in the rest frame.
The wavelength units are the same as those declared in the target
lime.Spectrumorlime.Cubeobservations.
Default bands database:
 includes a database with common lines which can be observed in astronomical spectra. To access this database you can use the
includes a database with common lines which can be observed in astronomical spectra. To access this database you can use the `lime.line_bands <https://lime-stable.readthedocs.io/en/latest/introduction/api.html#lime.line_bands>`__ function:
[2]:
import numpy as np
from astropy.io import fits
from IPython.display import Image, display
from pathlib import Path
import lime
bands_df = lime.line_bands()
The default table wavelengths are in angstroms with the observed wavelength and band boundaries (w1, w2,w3,w4,w5,w6) values in air for the  <
< 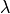 <
< 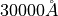 interval
interval
However, you can constrain the output bands from the `lime.line_bands <https://lime-stable.readthedocs.io/en/latest/introduction/api.html#lime.line_bands>`__ function using its attributes. For example, you can limit the output line bands by a wavelenght interval with wave_inter, as well as a lines_list and particle_list. Regarding the output values, you can specify the units_wave and whether to output the vacuum wavelengths via vacuum_conversion=True. Finally, you can
state the number of decimals on the line labels using the sig_fig attribute:
[3]:
lime.line_bands(wave_intvl=(300, 900), particle_list=('He1','O3','S2'), units_wave='nm', decimals=None, vacuum=True)
[3]:
| wavelength | wave_vac | w1 | w2 | w3 | w4 | w5 | w6 | latex_label | units_wave | particle | transition | rel_int | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| He1_403nm | 402.733452 | 402.73345 | 401.503486 | 402.244991 | 402.270129 | 403.335171 | 403.435647 | 404.138977 | He1-$403nm$ | Angstrom | He1 | rec | 0 |
| S2_407nm | 406.974903 | 406.97490 | 405.938106 | 406.587703 | 406.630488 | 407.441240 | 408.159187 | 408.819158 | S2-$407nm$ | Angstrom | S2 | col | 0 |
| O3_436nm | 436.443598 | 436.44360 | 429.897570 | 432.494291 | 435.817822 | 437.401787 | 439.206155 | 441.750863 | O3-$436nm$ | Angstrom | O3 | col | 0 |
| He1_447nm | 447.274040 | 447.27404 | 445.323193 | 446.639157 | 446.851915 | 447.876791 | 448.209876 | 449.824056 | He1-$447nm$ | Angstrom | He1 | rec | 0 |
| He1_492nm | 492.330504 | 492.33050 | 490.718146 | 491.762466 | 491.908118 | 492.953284 | 493.204228 | 494.116484 | He1-$492nm$ | Angstrom | He1 | rec | 0 |
| O3_496nm | 496.029500 | 496.02950 | 493.073376 | 494.818947 | 494.976792 | 497.217554 | 497.428449 | 498.588373 | O3-$496nm$ | Angstrom | O3 | col | 0 |
| O3_501nm | 500.824004 | 500.82400 | 497.326052 | 498.598165 | 499.681938 | 502.578172 | 502.922278 | 504.528111 | O3-$501nm$ | Angstrom | O3 | col | 0 |
| He1_588nm | 587.724329 | 587.72433 | 584.742570 | 586.742789 | 586.886434 | 588.729942 | 589.010016 | 590.358048 | He1-$588nm$ | Angstrom | He1 | rec | 0 |
| He1_668nm | 667.999556 | 667.99955 | 666.105098 | 667.204538 | 667.267813 | 669.001580 | 669.174956 | 670.012944 | He1-$668nm$ | Angstrom | He1 | rec | 0 |
| S2_672nm | 671.829502 | 671.82950 | 668.873329 | 670.826383 | 670.941255 | 672.742554 | 674.640249 | 676.192505 | S2-$672nm$ | Angstrom | S2 | col | 0 |
| S2_673nm | 673.267400 | 673.26740 | 668.873329 | 670.826383 | 672.741766 | 674.343008 | 674.640249 | 676.192505 | S2-$673nm$ | Angstrom | S2 | col | 0 |
| He1_707nm | 706.716328 | 706.71633 | 703.114524 | 704.368881 | 705.727267 | 708.149955 | 708.955184 | 710.651666 | He1-$707nm$ | Angstrom | He1 | rec | 0 |
Using a dataframe:
In  , a bands table (and the output line measurement tables) variables are pandas Dataframea.
, a bands table (and the output line measurement tables) variables are pandas Dataframea.
To get the data from a certain column you can use several commands:
[4]:
# Table columns
print('Columns', bands_df.columns)
# The index is the first column which is used to index the columns:
labels = bands_df.index.to_numpy()
# Get certain columns
ions = bands_df['particle'].to_numpy()
wave_array = bands_df.wavelength.to_numpy()
# First five values from these columns
print(f"{labels[:5]}, {ions[:5]}, {wave_array[:5]}")
Columns Index(['wavelength', 'wave_vac', 'w1', 'w2', 'w3', 'w4', 'w5', 'w6',
'latex_label', 'units_wave', 'particle', 'transition', 'rel_int'],
dtype='object')
['H1_1215A' 'C4_1548A' 'He2_1640A' 'O3_1666A' 'C3_1908A'], ['H1' 'C4' 'He2' 'O3' 'C3'], [1215.1108 1547.6001 1639.7896 1665.5438 1908.0803]
Similarly, you can use these comands to get the data from the rows:
[5]:
H1_1215A_params = bands_df.iloc[0].to_numpy()
H1_4861A_params = bands_df.loc['H1_4861A'].to_numpy()
print(H1_1215A_params)
print(H1_4861A_params)
[1215.1108 1215.6699 1100.0 1150.0 1195.0 1230.0 1250.0 1300.0
'H1-$1215\\mathring{A}$' 'Angstrom' 'H1' 'rec' 0]
[4861.2582 4862.691 4809.8 4836.1 4848.715437 4876.181741 4883.13 4908.4
'H1-$4861\\mathring{A}$' 'Angstrom' 'H1' 'rec' 0]
Finally, you can combine these commands to access the data from certain cells:
[6]:
bands_df.at['H1_1215A', 'wavelength']
[6]:
1215.1108
[7]:
bands_df.loc['H1_1215A', 'wavelength'], bands_df.loc['H1_1215A'].wavelength
[7]:
(1215.1108, 1215.1108)
[8]:
bands_df.loc[['H1_1215A', 'H1_4861A'], 'wavelength'].to_numpy(),
[8]:
(array([1215.1108, 4861.2582]),)
[9]:
bands_df.loc['H1_1215A':'He2_1640A', 'wavelength'].to_numpy()
[9]:
array([1215.1108, 1547.6001, 1639.7896])
[10]:
bands_df.loc['H1_1215A', 'w1':'w6'].to_numpy()
[10]:
array([1100.0, 1150.0, 1195.0, 1230.0, 1250.0, 1300.0], dtype=object)
Load/save a bands dataframe/measurements log:
To save these dataframes you can use the `save_frame <https://lime-stable.readthedocs.io/en/latest/introduction/api.html#lime.save_log>`__:
[11]:
# Save to the current folder in several formats:
lime.save_frame('bands_frame.txt', bands_df)
lime.save_frame('bands_frame.pdf', bands_df, parameters=['wavelength', 'latex_label'])
In files with a multi-page structure you can specify the extension:
[12]:
lime.save_frame('bands_frame.xlsx', bands_df, page='BANDS')
lime.save_frame('bands_frame.fits', bands_df, page='BANDS')
Similarly, to read these files as a pandas dataframe you can use:
[15]:
bands_df_txt = lime.load_frame('bands_frame.txt')
bands_df_xlsx = lime.load_frame('bands_frame.xlsx', page='BANDS')
bands_df_fits = lime.load_frame('bands_frame.fits', page='BANDS')
Updating the bands database:
The user is adviced to keep his/her own bands database with the bands and latex labels he/she prefers. Afterwards, you can use it in  functions (including
functions (including lime.line_bands) to run your scripts. However, in case you need to update the default database you can recover its location with this command:
[17]:
lime.io._parent_bands_file
[17]:
PosixPath('/home/vital/PycharmProjects/lime/src/lime/resources/parent_bands.txt')
Please remember: If you need to update the default  lines database make sure to fill all the columns. If there is any issue you can download the default database from the github (or reinstall
lines database make sure to fill all the columns. If there is any issue you can download the default database from the github (or reinstall  )
)